The ROKS research
group aims to collect and share knowledge
and experience in medical physics and
radiation oncology in the form of electronic
data. The objectives of the group are:
(a) develop
knowledge-based and evidence-based
radiotherapy treatments,
(b) use data to
improve the experience and outcomes of
radiation oncology patients.

Our research group ROKS!
(left to right: John
Kildea, Ackeem Joseph, Logan Montgomery,
David Hererra)
Humans are gifted with a great capacity
to learn. We learn from our experiences and
mistakes, and from the experiences and
mistakes of others. For centuries we have
shared knowledge using spoken word and
hand-written texts. Today, we have easy access
to vasts amounts of knowledge in digital
format. We can efficiently share our knowledge
and experience electronically and we can
create new knowledge by collecting and
examining data. Medicine is no exception. The
medical field produces vast amounts of data,
making it ripe for the electronic collection
and sharing of knowledge and experience.
The ROKS research group aims to collect
and share knowledge and experience in medical
physics and radiation oncology using three
methods:
1. by facilitating
guidelines, policies and procedures written by
experts,
2. by learning from adverse events, and
3. by learning from data.
The ROKS research group currently
comprises one PI (John Kildea),
two M.Sc students (Ackeem Joseph and Logan
Montgomery) and one research assistant (David
Herrera). Our group is an integral part of the
HIG (Health Informatics Group).
Our current research projects include:
- Realistic
knowledge-based waiting times for
Radiation Oncology patients (the HIG)
- Depdocs
(Ackeem Joseph and John Kildea)
- SaILS NSIR-RT
(Logan Montgomery and John Kildea)
- AEHRA (Ackeem
Joseph and John Kildea)
- DVH registry
(Ackeem Joseph, John Kildea and Dr. Carolyn
Freeman, MD)
- ATS (Dr. Tarek
Hijal, MD, Ackeem Joseph, John Kildea)
Each of these research projects is
described in detail below.
The Health
Informatics Group Collaboration
 Some members of the Health Informatics
Group, November 2015.
Some members of the Health Informatics
Group, November 2015.
In 2014 John Kildea (medical physicist), Laurie
Hendren (computer scientist) and Tarek
Hijal (radiation oncologist) formed the Health
Informatics Group collaboration. Over the
past 1.5 years the collaboration's translational
research has involved a total of 14 student
researchers and has given rise to Opal, the
oncology portal and application. When released
in the Spring of 2016, Opal will provide
radiation oncology patients with access to their
medical data and to real-time estimates of their
wait times, determined by applying
machine-learning algorithms to the time-stamp
data of previous patients.
In October 2014, the HIG received its first
research grant, $150k from the MUHC Q+
initiative, for our research project entitled "Realistic
knowledge-based waiting times for Radiation
Oncology patients - addressing the pain of
waiting". For more details about the
HIG, please see our webpage.
Project 1:
Realistic knowledge-based waiting times for
Radiation Oncology patients
PIs: Laurie
Hendren, Tarek Hijal, John Kildea (co PIs)
Current Students:
- Ackeem Joseph, B.Sc, M.Sc
candidate, Medical Physics
- David Hererra, B.Sc, Senior
developer
- Faiz Khan, B.Sc, M.Sc candidate,
Computer Science
- Simon Labute, B.Sc. student,
Volunteer developer
- Claudine Lebosquain, B.Sc,
(Computer Science and Biology), Volunteer
developer
- Cloe Pou-Prom, B.Sc, Computer
Science and Biology
- Zachariah Stevenson, B.Sc,
student, Computer Science
- Russell Wang, B.Sc, student,
Computer Science and Biology
Past Students:
- Maxim Gorshkov, B.Sc student,
Computer Science
- Mehryar Keshavarz, B.Sc student
- Alvin Leung, B.Sc student
- Marya Sawaf, B.Sc student
- Justin Wainberg, B.Sc student,
Anatomy and Cell Biology
- Yi Fan Zhou, B.Sc student,
Computer Science
Funding: MUHC
Q+ Initiative (The Montreal General Hospital
Foundation and the MUHC). The medical physics
students working on this project are supported
in part by MPRTN/CREATE.
Description:
In this HIG research project we are
aiming to provide radiation oncology patients
with realistic estimates of their waiting
times. To do so, we are applying machine
learning algorithms to the time-stamp data of
patients previously seen for radiation
oncology treatments at the McGill University
Health Centre. Radiation oncology patients
typically experience three distinct waiting
times. They are:
1. Waiting in the waiting room
to see a physician (minutes to hours)
2. Waiting at home for the
treatment plan to be prepared (days to
weeks)
3. Waiting in the waiting room
for daily treatment (minutes to hours)
Our project is not attempting (at least
not initially) to decrease these waiting
times. Rather, our goal is to provide patients
with realistic estimates of how long they
should expect to wait. By doing so, we aim to
reduce waiting time anxiety and improve the
experience of radiation oncology patients.
Our waiting time project was recently
(January 2016) highlighted
on the homepage of the McGill University
Health Centre
The following presentations describe
our waiting time project:
- Presentation
by the HIG at the
Accreditation Canada Quality Conference,
Montreal, April 2016
- Presentation
by Hendren, Hijal
and Kildea, Board Meeting of the McGill
University Health Centre, April 2016
- Presentation
by Hendren, Hijal
and Kildea, Board Meeting of the MGH
Corporation, January 2016
- Presentation
by Hendren, Hijal
and Kildea, Patient Safety Week, October
2015
- Presentation
by Hendren, Hijal and Kildea, MUHC
Oncology Grand Rounds, March 2015
The following presentations and reports
describe the methods and results of our
current machine learning research for waiting
times. This effort is the focus of Ackeem
Joseph's M.Sc research project.
To communicate waiting time estimates
with patients we have developed two software
products:
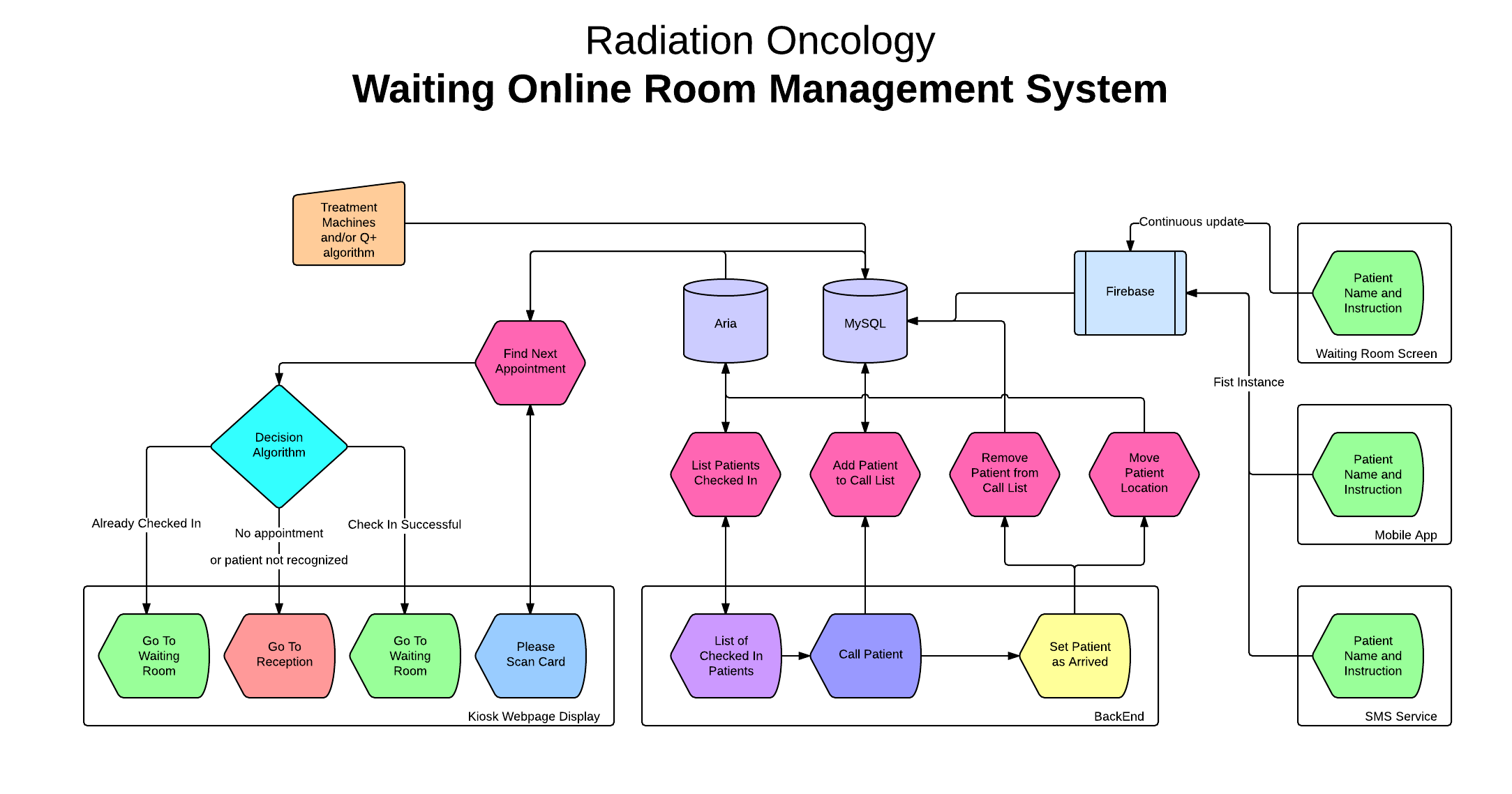
WORMS
(Waiting Online Room Management System) is a
software platform, written in AngularJS and
Firebase, to manage patient experience in the
waiting room. It comprises a user-friendly
kiosk check-in system, a virtual waiting room
whereby staff can monitor which patients are
waiting and for how long, and a digital screen
call-in system. From a research perspective,
WORMS provides the HIG team with valuable
time-stamp data regarding each patient's
trajectory. These data feed our machine
learning algorithm and will ultimately inform
future efficiency improvements. A technical
document describing WORMS is under
development.
Overview of the WORMS
data flow architecture.
WORMS is currently in use in the Cedars
Cancer Centre at the MUHC for radiotherapy and
chemotherapy patients. A possible extension of
WORMS to all ambulatory clinics of the MUHC is
under discussion (March, 2016).

Kiosk and screens in the Radiation
Oncology waiting room at the MUHC
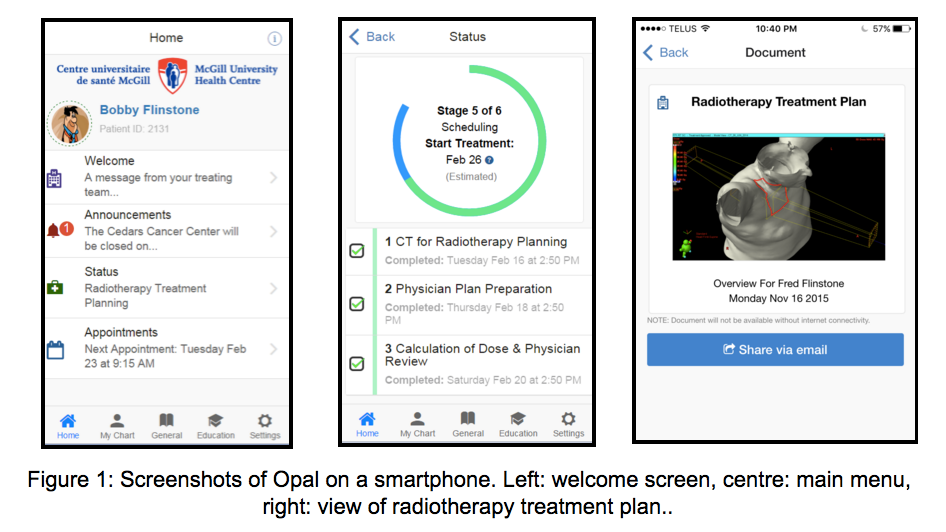
Opal
(Oncology portal and application) is a mobile
phone app and web portal for oncology
patients. When released
in the Spring of 2016, Opal will provide
radiation oncology patients with access to
their medical data and to real-time estimates
of their wait times, determined by applying
machine-learning algorithms to the time-stamp
data of previous patients. We are presently
recruiting patients for two Opal focus groups
in January 2016.
A technical document describing Opal is
available here.
New: Opal wins the Onsen
Ninja App Contest (February,
2016)!
Ultimately, we hope that Opal will also
provide the radiation oncology team with
valuable patient-reported outcome (PRO) data
in discrete format. PRO data from consenting
patients will be linked with dosimetric data (DVH registry) to aid
comparative effectiveness research. This
research project will be conceptually similar
to the Oncospace
project at Johns Hopkins and the popular patientslikeme.com
crowd-sourcing web site.
Project 2:
Depdocs
PI: John Kildea
Students: Ackeem Joseph, B.Sc (M.Sc
candidate), Ali Snan, B.Sc student (volunteer)
Funding: Ackeem
Joseph was funded through MPRTN/CREATE
Depdocs
is an information-sharing platform that was
designed to share healthcare knowledge by
facilitating collaboratively-written policies
and procedures. Depdocs was developed on the Drupal
framework. At the MUHC, Depdocs is housed inside
the Department of Medical Physics but accessible
throughout the hospital network. Currently over
150 users access over 700 documents within the
system. 35 committees use Depdocs to manage
their private documents and schedules and over
300 problem reports and change logs have been
recorded using the system over two years of
operation.
New: In
January 2016 Depdocs will be used for document
management by the Cancer Research Program of the
MUHC
Research Institute.
Depdocs was designed and developed by Ackeem
Joseph and John Kildea. A public version of the
platform is available at depdocs.com.
The following technical documents and
presentations describe the system:
We are currently working with McGill
undergraduate research student Ali Snan to gather usage and
user-feedback data for a peer-reviewed
publication in February 2015.
Project 3: SaILS
NSIR-RT
PI: John Kildea
(collaboration with Crystal Angers at the
Ottawa Hospital Cancer Centre)
Students: Logan Montgomery, B.Sc, M.Sc
candidate
Funding: Logan
Montgomery is supported by the CPQR and MPRTN/CREATE
"Always
make new mistakes" – Esther Dyson
SaILS
(Safety and Incident Learning System) is a
web-based platform to share and learn from
incidents in radiation oncology. SaILS was
originally developed by Randle Taylor
at the Ottawa Hospital Cancer Centre. McGill
medical physics M.Sc. student Logan Montgomery
has been leading the effort at the MUHC to
redevelop SaILS to include the incident
categorization taxonomy of NSIR-RT
(the National System for Incident Reporting -
Radiation Treatment) that was developed by CIHI in
collaboration with the CPQR.
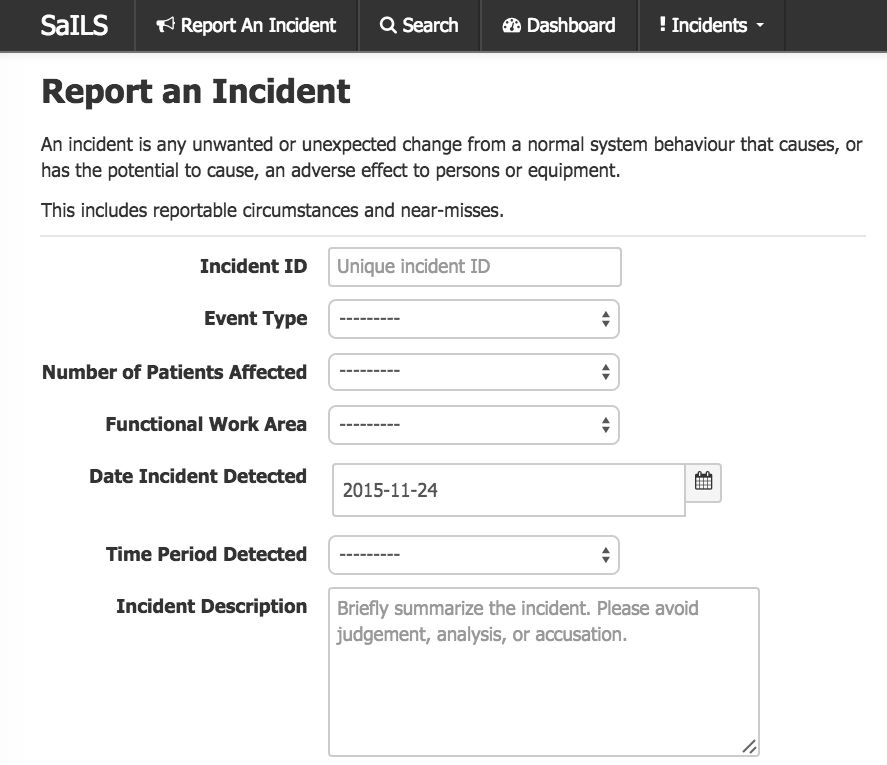
Screenshot of SaILS NSIR-RT at the MUHC.
SaILS NSIR-RT is now in operation at the MUHC.
An initial evaluation of the system will be
presented at the Canadian
Winter School for Quality and Safety in
Radiation Oncology in February 2016.
The following presentations describe Logan
Montgomery's research project:
Top
Project 4: AEHRA
PI: John Kildea
Students: Ackeem Joseph, B.Sc (M.Sc
candidate)
Funding: Ackeem
Joseph was supported by a studentship from
the Canadian
Patient Safety Institute and by MPRTN/CREATE.
AEHRA stands for Automated Electronic
Health Record Auditing. It is a software
infrastructure that is designed to automatically
run scheduled audits of patient electronic
health records to search for and flag outlier
data that may indicate errors or abnormalities.
During his CPSI studentship, Ackeem Joseph
developed the following:
- A data selection and aliasing
interface. Aliasing allows data that mean
the same thing to be labelled under one
umbrella reference/name,
- A data selection daemon
to copy relevant (and anonymized) medical
record data to a dedicated treatment
registry database, and
- A data analysis and visualization
webpage to search for outliers in medical
record data.
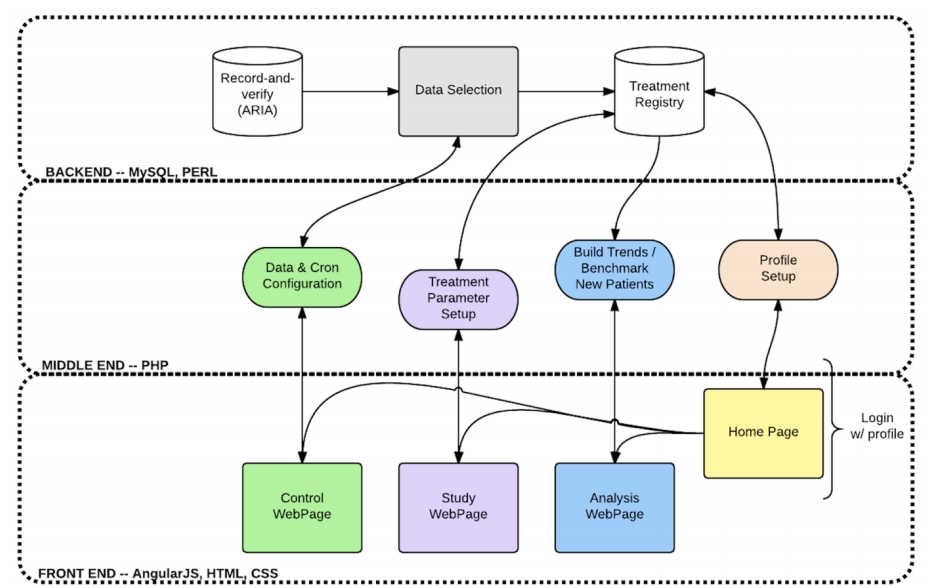 AEHRA infrastructure
AEHRA infrastructure
AEHRA is not yet in routine clinical
operation pending an improvement in its
ability to flag outlier data. However, the
software infrastructure that was developed to
select and copy AEHRA data has been cloned by
our team for three unrelated software projects
(ATS, Opal
and our machine learning project to estimate
waiting times).
The following reports and presentations
describe AEHRA in greater detail:
Top
Project5: DVH
Registry
PIs: John Kildea and Dr.
Carolyn Freeman, MD
Students: Ackeem Joseph, B.Sc (M.Sc
candidate), Jeremy
Ahearn, B.Sc student
Funding: Ackeem
Joseph and Jeremy Ahearn were supported by MPRTN/CREATE
The McGill DVH Registry is a software tool
to facilitate research and knowledge-based
treatment planning in radiation oncology. It
allows planners to store and retrieve in
aggregate the dose volume histograms of the
treatment plans that they or their colleagues
have prepared.
The registry records and summarizes data (ages,
sex, diagnosis, treatment site, etc) from the
plans of previously-treated patients and allows
for comparison of a present DVH with the
appropriate historical average. The backbone of
the registry is a MySQL database that stores DVH
data exported from the Eclipse treatment
planning software (Varian Medical Systems, Palo
Alto, CA); the front-end provides web pages via
a web-server. Before submission to the database,
each patient's data are anonymized and
structures are renamed/aliased using a naming
convention. The registry allows for addition of
future plans as they are produced, such that the
data set is always current.
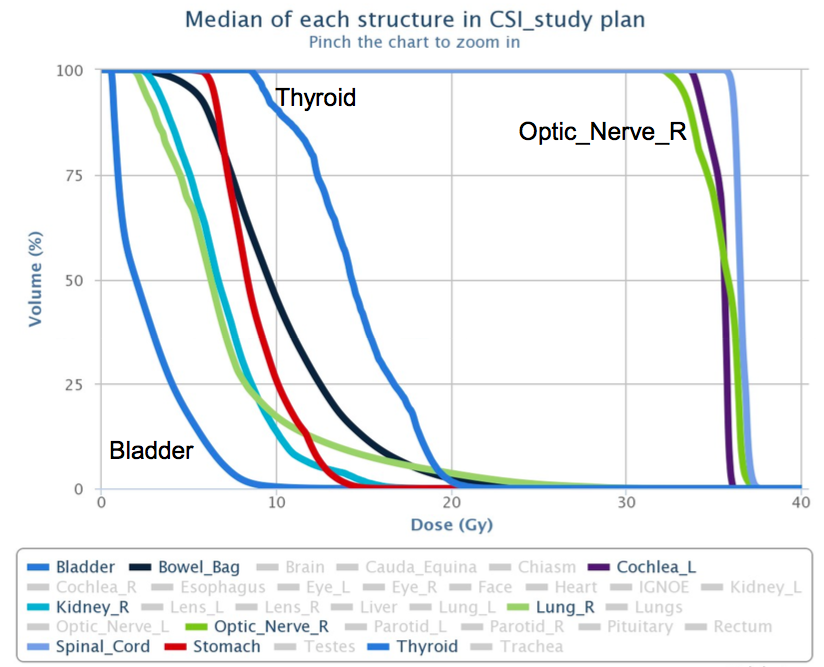
Screenshot from the DVH registry showing
aggregate median DVHs for CSI treatment plans.
The following presentations and reports describe
the DVH registry. A paper describing its use for
CSI planning at the MUHC is in preparation.
Top
Project5:
ATS (Aria to Streamline)
PIs: Dr. Tarek
Hijal, MD, John Kildea
Students: Ackeem Joseph, B.Sc
(M.Sc candidate)
Funding: Internal
MUHC support
Aria to Streamline is
an interface to send clinical documents from
the Aria information system (Varian Medical
Systems Inc.) in Radiation Oncology to Oacis
(Telus Health Inc.), the MUHC's electronic
medical record system. The interface was
developed with student Ackeem Joseph and is
based on the architecture that we put in
place for the AEHRA project.
Using a 5-minute cron SQL query in linux,
word documents, created in Aria, are located
in the Aria database, converted into pdf
format using the open source Libreoffice
command line tool lowriter, and then
send to the ftp input of Streamline
Health's Access Anyware gateway.
Access Anyware is, in turn, used by Oacis to
display clinical documents.
Architecture of
the ATS interface.
ATS is in routine clinical operation and has
been used to transfer over three thousand
clinical documents in one year of operation.
A manuscript detailing the project is in
preparation.
Top
|